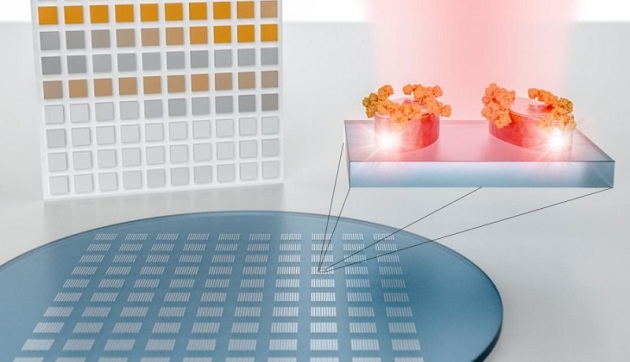
A new system can detect and analyze molecules without the need for an infrared spectrometer. The system uses nanostructured metapixels to detect and then translate molecules’ unique signatures into bar codes. The technology can be integrated into a compact sensor chip. It opens the door to large-scale image-based detection of materials using machine-learning technology.
Infrared spectroscopy is the benchmark method for detecting and analyzing organic compounds. But it requires complicated procedures and large, expensive instruments, making device miniaturization challenging and hindering its use for some industrial and medical applications and for data collection out in the field, such as for measuring pollutant concentrations. Furthermore, it is fundamentally limited by low sensitivities and therefore requires large sample amounts.
However, scientists at EPFL’s School of Engineering and at Australian National University (ANU) have developed a compact and sensitive nanophotonic system that can identify a molecule’s absorption characteristics without using conventional spectrometry.
Their system consists of an engineered surface covered with hundreds of tiny sensors called metapixels, which can generate a distinct bar code for every molecule that the surface comes into contact with. These bar codes can be massively analyzed and classified using advanced pattern recognition and sorting technology such as artificial neural networks.
Translating molecules into bar codes
The chemical bonds in organic molecules each have a specific orientation and vibrational mode. That means every molecule has a set of characteristic energy levels, which are commonly located in the mid-infrared range — corresponding to wavelengths of around 4 to 10 microns. Therefore, each type of molecule absorbs light at different frequencies, giving each one a unique “signature.” Infrared spectroscopy detects whether a given molecule is present in a sample by seeing if the sample absorbs light rays at the molecule’s signature frequencies. However, such analyses require lab instruments with a hefty size and price tag.
The pioneering system developed by the EPFL scientists is both highly sensitive and capable of being miniaturized; it uses nanostructures that can trap light on the nanoscale and thereby provide very high detection levels for samples on the surface. “The molecules we want to detect are nanometric in scale, so bridging this size gap is an essential step,” says Hatice Altug, head of EPFL’s BioNanoPhotonic Systems Laboratory and a coauthor of the study.
The system’s nanostructures are grouped into what are called metapixels so that each one resonates at a different frequency. When a molecule comes into contact with the surface, the way the molecule absorbs light changes the behavior of all the metapixels it touches.
“Importantly, the metapixels are arranged in such a way that different vibrational frequencies are mapped to different areas on the surface,” says Andreas Tittl, lead author of the study.
This creates a pixelated map of light absorption that can be translated into a molecular bar code — all without using a spectrometer.
The scientists have already used their system to detect polymers, pesticides and organic compounds. What’s more, their system is compatible with CMOS technology.
“Thanks to our sensors’ unique optical properties, we can generate bar codes even with broadband light sources and detectors,” says Aleksandrs Leitis, a coauthor of the study.
There are a number of potential applications for this new system. “For instance, it could be used to make portable medical testing devices that generate bar codes for each of the biomarkers found in a blood sample,” says Dragomir Neshev, another coauthor of the study.
Artificial intelligence could be used in conjunction with this new technology to create and process a whole library of molecular bar codes for compounds ranging from protein and DNA to pesticides and polymers. That would give researchers a new tool for quickly and accurately spotting miniscule amounts of compounds present in complex samples.